Research
Our long-term research interest is to decode the human genome. The recent research focus of my lab is on developing and applying computational and high-throughput experimental methods to understand the gene regulatory roles of non-coding genetic variants in different pathological conditions, including cancers and neurodegenerative diseases. Specifically, we focused on the computational method development for circulating cell-free DNA fragmentation and new biotechnology development for the joint profiling of multi-omics within the same single cells, which will eventually enable biomarker discovery for the early diagnosis and prognosis of many complex diseases.
Current Projects
Artificial intelligence for liquid biopsy
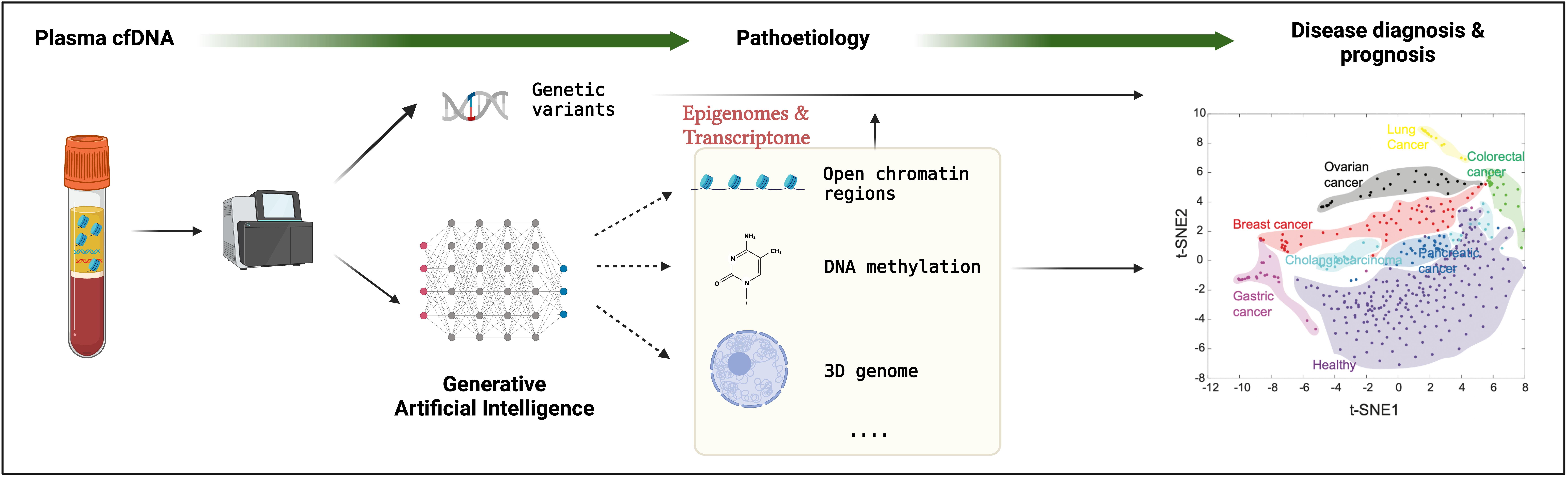
Circulating cell-free DNA (cfDNA) is released into the peripheral blood after cellular death and recycled every few minutes for up to 12 hours. The collection of cfDNA fragments represents a real-time in vivo snapshot of the genome from cells contributing to cfDNA. Interestingly, cfDNA is not randomly fragmented and has recently been associated with the epigenetic status within the cells. This suggests the possibility of computationally inferring the cellular epigenomes from cfDNA fragmentation. However, computational methods have yet to be developed to construct cellular epigenomes from cfDNA assays. Millions of cfDNA genomic sequencing datasets are generated in clinics every year. To leverage these cfDNA datasets and advance our understanding of the epigenomes and, thus, the function of non-coding regulatory elements, we are developing computational methods based on generative artificial intelligence approaches to reconstruct the cellular epigenomes and transcriptomes from a single cfDNA assay. Finally, we can identify meaningful biomarkers from cfDNA for the early diagnosis and prognosis of different cancers and many other complex diseases, such as neurodegenerative and neuroinflammation diseases.
Single-cell & single-molecule multi-omic technology to decode genome
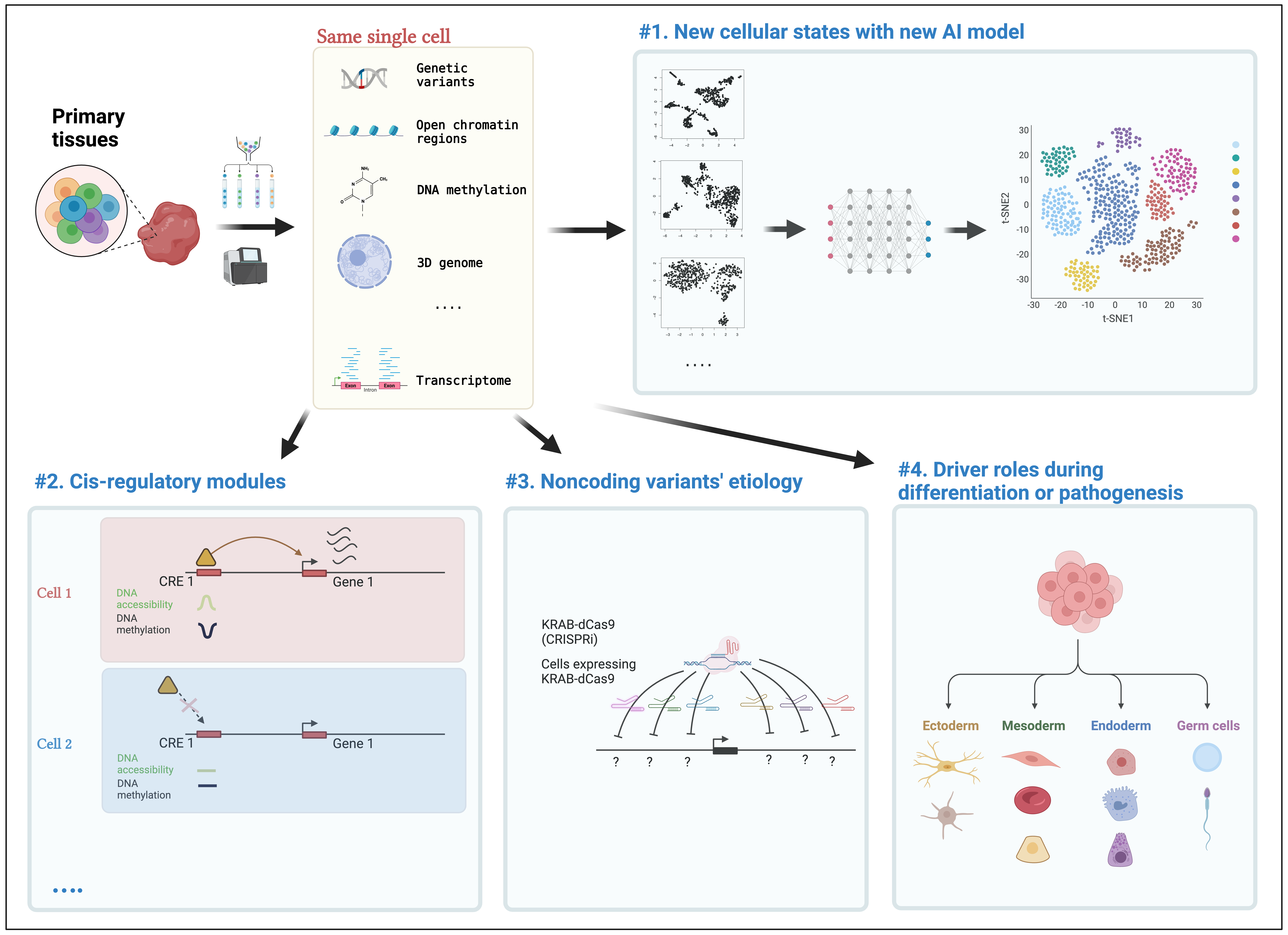
Epigenetic modifications, including DNA methylation, histone modifications, and three-dimensional (3D) genome topology, combine with genetic content to determine the mammalian transcriptional factor (TF) binding and, thus, gene regulation. However, gene activation or repression potential cannot be entirely predicted by looking at a single molecular measurement. Accurate predictive models require multiple molecular measurements simultaneously. We are currently limited by the number of simultaneous measurements we can perform in a single cell. In addition, the interactions between different epigenetic marks and their effects on gene expression are currently studied either in homogenous cultured cells or bulk tissues that average the readout. The study of interactions between different cell-type-specific epigenetic marks and gene expression in heterogeneous tissues at the single cell level is still in its infancy. We have developed several technologies to simultaneously capture multiple molecular measurements in the same assay or single cells. We will continue to develop more powerful multi-omic technologies to dissect the regulatory roles of the non-coding regulatory elements in the genome and finally identify the therapeutic targets for human diseases.
Funding
Our work is currently (and was previously) supported by NIH (R35GM147283, R56HG012360), NSF (ACCESS and XSEDE Resource Allocation Service), Bill & Melinda Gates Foundation (MOMI Ideas Award), Sony Research Foundation (Sony Focus Research Award), and Northwestern University (Yaping Liu’s start-up).