Tools
Featured
 FinaleMePredict cfDNA methylation and its tissues-of-origin by cfDNA fragmentation patterns
FinaleMePredict cfDNA methylation and its tissues-of-origin by cfDNA fragmentation patternsFinaleMe (FragmentatIoN AnaLysis of cEll-free DNA Methylation) is a Java program to predict DNA methylation in deep and low-coverage cell-free DNA WGS data without other training data.
 FinaleDBA browser and database of cell-free DNA fragmentation patterns
FinaleDBA browser and database of cell-free DNA fragmentation patternsWe collected thousands of uniformly processed and curated de-identified cfDNA WGS datasets across different pathological conditions. FinaleDB comes with a fragmentation genome browser, from which users can seamlessly integrate thousands of other omics data in different cell types to experience a comprehensive view of both gene-regulatory landscape and cfDNA fragmentation patterns.
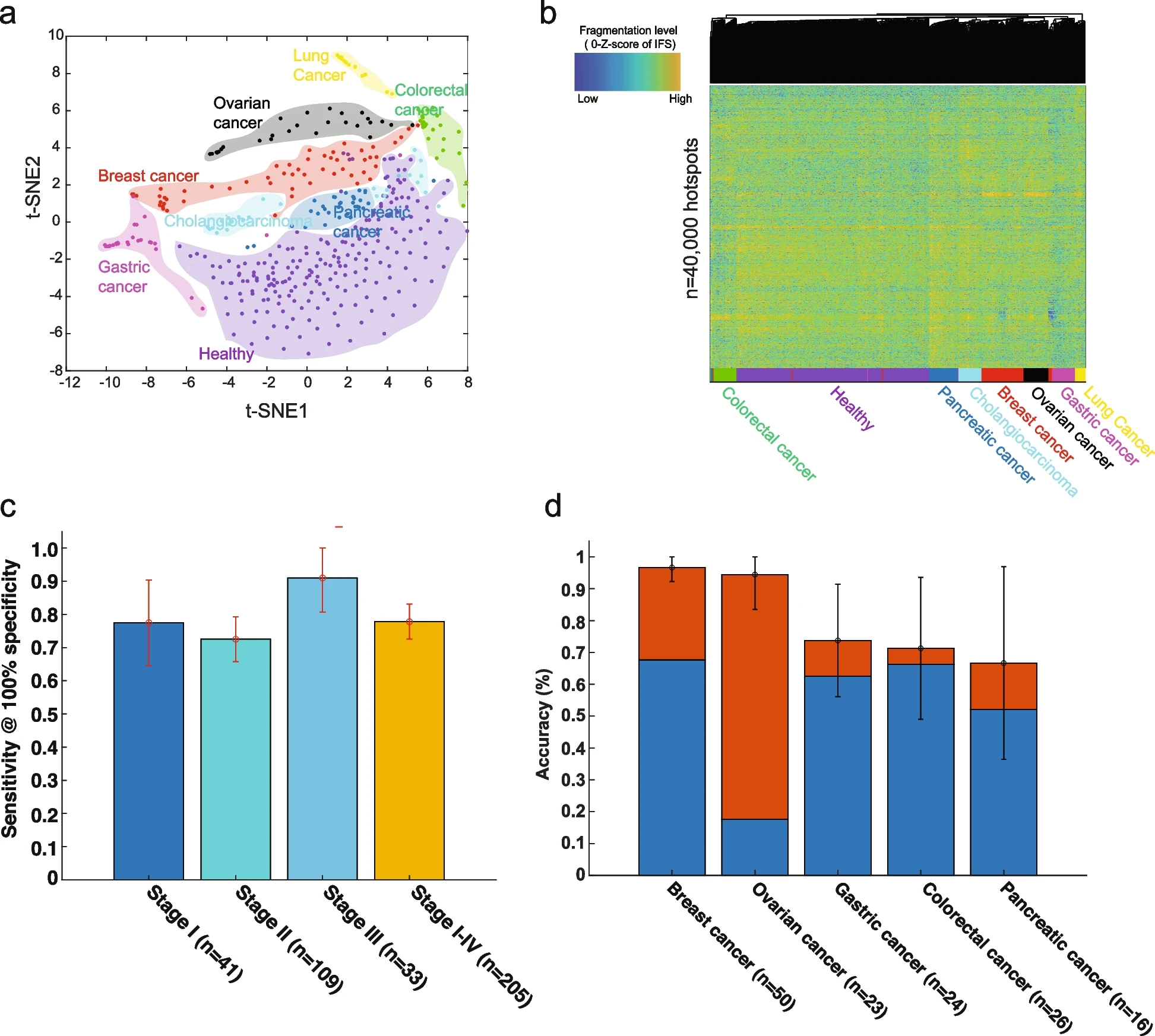 CRAGDe novo characterization of cell-free DNA fragmentation hotspots
CRAGDe novo characterization of cell-free DNA fragmentation hotspotsWe developed an approach to de novo characterize the cfDNA fragmentation hotspots from whole-genome sequencing. In healthy, cfDNA fragmentation hotspots are highly enriched in gene-regulatory elements, especially open chromatin regions from hematopoietic cells. The results highlight the significance of de novo characterizing the cell-free DNA fragmentation hotspots for detecting early-stage cancers and dissection of gene-regulatory aberrations in cancers.
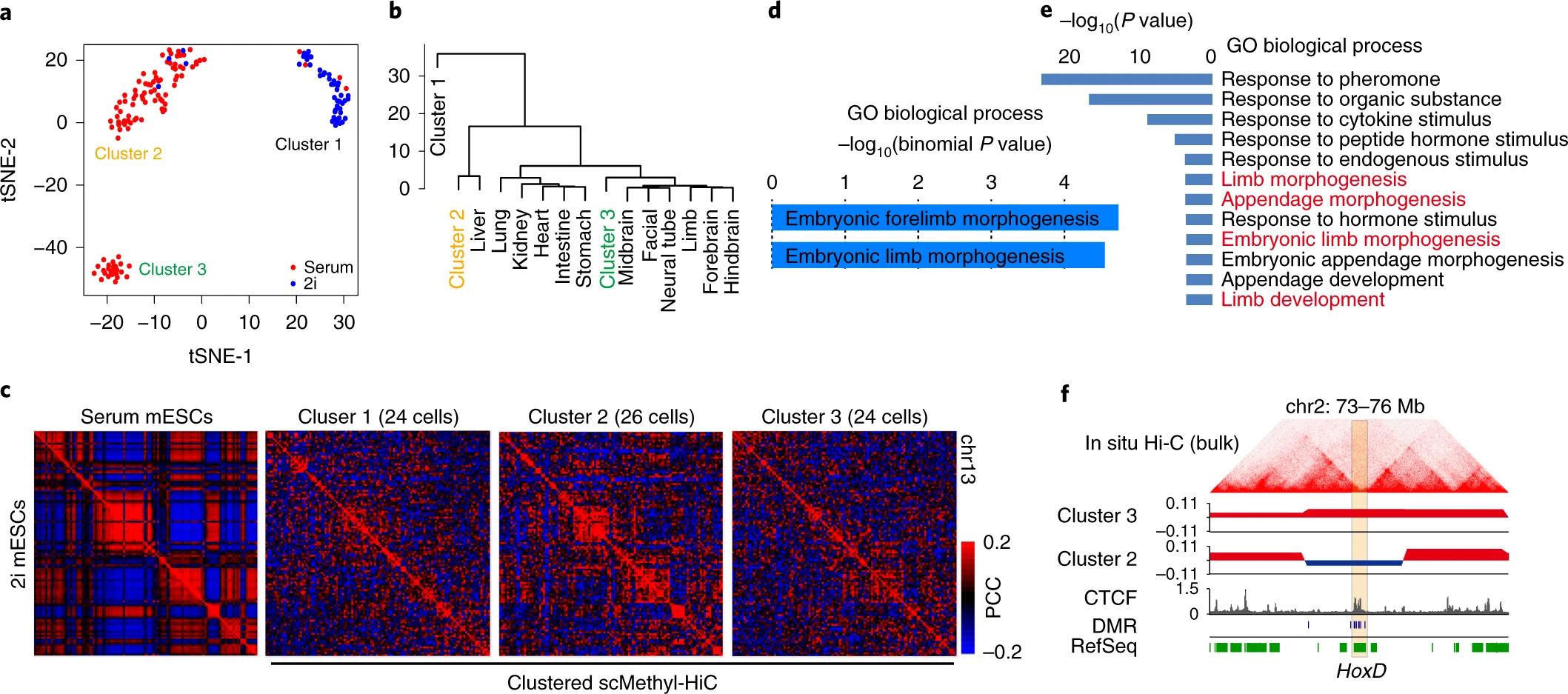 BhmemComputational tools for Methyl-HiC (Li, Liu et al. 2019 Nature Methods) and NOMe-HiC (Fu et al. 2023 Genome Biology)
BhmemComputational tools for Methyl-HiC (Li, Liu et al. 2019 Nature Methods) and NOMe-HiC (Fu et al. 2023 Genome Biology)Hi-C reads at bisulfite converted space are hard to be mapped. Bhmem is based on bwa aligner but aware of bisulfite converted reads and genome.
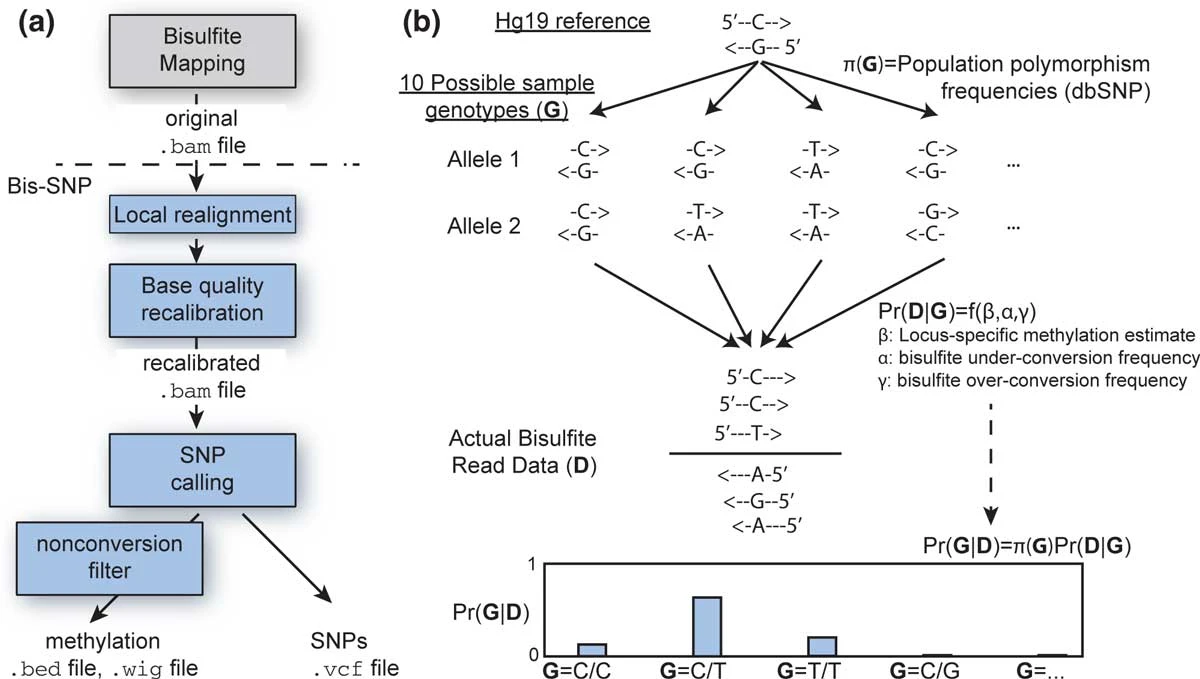 Bis-SNPCombined SNP and methylation caller at bisulfite sequencing
Bis-SNPCombined SNP and methylation caller at bisulfite sequencingBisSNP is a package based on the Genome Analysis Toolkit (GATK) map-reduce framework for genotyping and accurate DNA methylation calling in bisulfite treated massively parallel sequencing (Bisulfite-seq, NOMe-seq, RRBS and any other bisulfite treated sequencing) with Illumina directional library protocol.